Quick start tutorial -- De novo drug design
Author: Qifeng Bai
SARS-CoV-2 caused the rapid spread of coronavirus disease 2019 (COVID-19) throughout the world. In this tutorial, the SARS-CoV-2 main protease (Mpro) which plays an important role on the replication of coronavirus is selected as the example target. The crystal structures of SARS-CoV-2 Mpro have been reported (PDB ID: 6LU7, 6Y2F, etc) [1,2]. Here, the built structure of SARS-CoV-2 Mpro provided by the group of Prof. Zihe Rao is selected as our quick start example (see Figure 1). Here, MolAICal (https://molaical.github.io or https://molaical.gitlab.io) is employed for this tutorial.
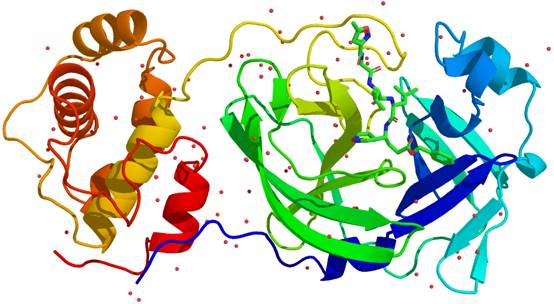
Figure 1. The structure of SARS-CoV-2 Mpro.
Materials
1. Software requirement
1) MolAICal: https://molaical.github.io
2) UCSF Chimera: https://www.cgl.ucsf.edu/chimera/
Make sure every software is installed rightly.
2. Example files
All the necessary tutorial files are downloaded from: QuickStart
Procedure
1. Open “InputParFile.dat” file, it needs to modify four parameters for this tutorial:
------------------------------------------------------------------------------------------
receptorPDB mproNolig.pdb
startFragFile startFrag.mol2
centerPoints -10.733 12.416 68.829
boxLengthXYZ 30.0 30.0 30.0
-------------------------------------------------------------------------------------------
The "receptorPDB" is set to the PDB format structure of SARS-CoV-2 Mpro without ligand. The "startFragFile" is set to the initial fragment. The "centerPoints" and "boxLengthXYZ" represent the box center and length, respectively.
2. Download and open tutorial folder "000-quickStart", then run command as below:
#> molaical.exe -denovo grow -i InputParFile.dat
Results
The calculation results are stored in the folder '001-AIGrow/results', while the minimum energy results are saved in '001-AIGrow/final_min_results'. The results in 'final_min_results' are generally considered better. Additionally, users may also perform the comprehensive analyses by considering the results in both 'results' and 'final_min_results' based on specific research requirements. The file "AstatisticsFile.dat" records the information of designed ligands that contain ID, Name, Cluster, Affinity, Formula, InChIKey, etc. The results are just a simple demo example that does not contain complete running results. The complete results can be obtained by finishing this task.
Open crystal ligand N3 named "ligand.mol2" of SARS-CoV-2 Mpro and generated ligand named "lig_11.mol2" (see Figure 2):
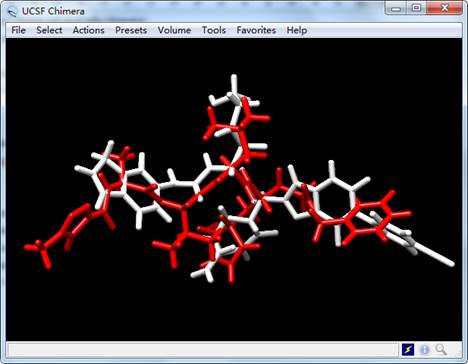
Figure 2. The results of generated ligand (white) and inhibitor N3 (red) of SARS-CoV-2 Mpro.
The detailed tutorial can be referred in part "1.1. Tutorials of 3D drug design by AI and de novo method" on https://molaical.github.io/tutorial.html.
References
[1] Jin, Z. et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature (2020).
[2] Zhang, L. et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science (2020).